05.01.2026
From Heatmap to Safety Assessment: Rapid Off-Target Screening in HLA-Compass
05.01.2026

How to search for offtargets in our HLACompass platform?
Today, I asked ChatGPT to convert the xscans heatmap of the paper of Johanna Olweus, that I posted yesterday into a csv matrix file.
After only 5 minutes and one refinement, it managed to extract the right PSSM matrix 👏
So I went to our HLA-Compass web interface at hla-compass.com and went to the toxicity screening tab, pasted the matrix from ChatGPT and voila, the results appeared.
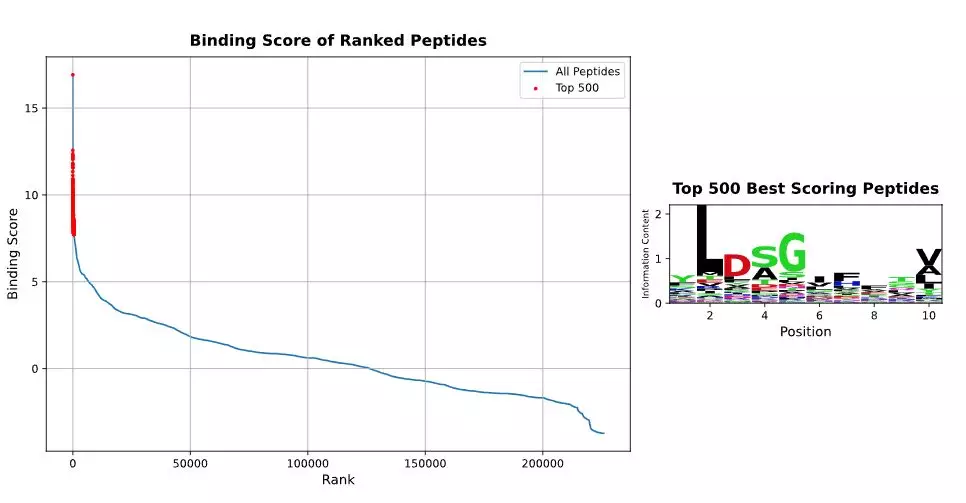
It indeed is a very specific TCR binding profile to HLA peptides. The top hit was the endogenous WT CTNNB1 epitope that was detected on THP1 cells. The second best peptide has already a much lower score. Some peptides rank still very similar as compared to the four peptides from the paper. One candidate, ALDSGAFQSV that is found on several healthy tissues (displayed in green) is predicted to be more similar to the target peptide as compared to the tested peptides.
So in 15 min max, I came from a published TCR where the x-scan needed to be extracted from a heatmap to potential risk candidates that can be tested in biologic assays. How long does this take your teams?
TCRtherapy immunotherapy cancertoxicologyimmunopeptidomics riskpeptides